Some editions of Lincoln Logs have specialized pieces for
building special buildings. These buildings have different purposes, like a
sawmill or a bank, and the specialized pieces help them carry out their
function of being that building.
Low and behold, there are special building blocks for building
specialized nucleic acid structures; usually these are RNAs for which the usual
building blocks just won’t do. These are the exceptions to the nucleotide rules
of A, C, G, and T for DNA and A C, G, and U for RNA.
There are a few different nucleotides located in DNA
molecules, but to date all these have been found to be damaged bases. Oxidized
guanosine bases have been the most commonly identified mutations, because
guanine is more susceptible to oxidation than the other bases. However, a recent study has identified a 6-oxothymidine in the placental DNA of a smoker.
More than 20 oxidized DNA bases have been found at one time
or another. Their importance lies in their inability to direct correct base
pairing in a replicating DNA or a transcribed RNA. In particular,
8-oxoguanosine in a DNA molecule often base
pairs with A instead of C, while an oxidized 8-oxoguanosine nucleotide (damaged
before it is incorporated into a DNA) will often be put in where a T should
rightfully have been placed.
Both of these problems would lead to mistakes in replication
or transcription. Some of these mistakes could be in places that matter. If
they change a codon, they might cause the wrong amino acid to be incorporated
and the resulting protein might be nonfunctional. Or they could create or
destroy a stop codon or a splice site. These would definitely alter the
resulting protein. Mistakes like this spell disease or cancer.
Oxoguanosine has been the most studied of the oxidized
bases, and several diseases have been linked to this mutation. Many cancers
have shown this mutation – leukemias, breast cancer, colorectal cancer, etc.
But in addition, things like Parkinson’s disease, Huntington’s disease, Lou
Gherig’s disease (ALS), and cystic fibrosis have been correlated with
8-oxoguanosine.
Don’t make the mistake of assuming that an 8-oxoguanosine is
the cause of any or all of these diseases, most have many potential causes. The
point is that this mutation may
contribute to these diseases in some cases. The point then is to find out how
to better prevent or repair them. However, your body is pretty good at doing
this itself – if everything is behaving normally.
There are specific repair pathways dedicated to removing and
replacing oxidized bases (base excision
repair or BER) or for nucleotides that
contain oxidized bases (nucleotide
excision repair or NER) in DNA. In
RNA, the major process to deal with 8-oxoguanosine is to destroy the damaged
RNA. There are actually several overlapping and redundant repair pathways for
8-oxoguanosine, suggesting that this mutation is particularly damaging and must
be dealt with for proper cell function.
It is when the body’s sensing and repair mechanisms don’t
work that the problems begin. Therefore, science needs to find better ways to
tell when the natural processes aren’t working and develop artificial ways to
reverse the damage. A 2013 review is showing the way to detecting mutated guanines in bodily fluids and tissues.
Specifically, this study looked at methods of detecting
8-oxoguanosine levels in plasma, urine, and cerebrospinal fluid and what those
changes might mean. The levels found represent a balance between the production
and repair of the mutations, so an increase means that more mistakes are being made, or
fewer are being repaired. Either way, it means that something must be done.
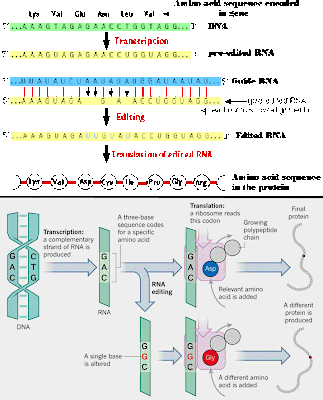
RNA editing takes place all the time, where RNA bases are changed after the RNA is transcribed from DNA.
In the majority of cases, the RNA editing modifies a standard nucleoside to
another standard nucleoside, or add/subtract nucleotides.
Insertion/deletion
edits for uracils can increase or decrease the length of the transcript.
The mRNA is paired with a guide RNA
(gRNA) and base-pairing takes place. For insertion, when there is a mismatch between the mRNA and the gRNA,
the editosome inserts a U, so the
mRNA transcript gets longer. In deletion editing, if there is an unpaired U in the
mRNA, it gets cut out, so the transcript gets shorter.
This was first discovered in a parasite called Trypanosoma brucei, the causative agent
of African Sleeping Sickness. There are so many positions at which these
insertions/deletions take place that it has come to be known as pan-editing.
In other cases, the editing takes the form of C being
replaced by a U. In some cases this results in a protein sequence different than that coded for by the
DNA - on purpose!! If that isn’t an exception, I don’t know what is. Other
times, the changing of a C to a U creates a stop codon.
In the human apolipoprotein B transcript, the intestinal
version undergoes the C to U editing and creates a stop codon, so the
apolipoprotein B is 48 kD in mass (B48). In the liver, no editing
takes place, so the protein is much larger (B100).
a 2013 database and algorithm to do nothing but predict C to U and U to C edits. Yes, there are U to C edits as
well, but only in plant mitochondria and plastids. As far as is known, U to C
edits work to destroy stop codons.
Then there is A to I editing. Wait you say, there’s no I in
nucleic acids (well, there are actually two “i”s, but you know what I mean). “I”
stands for inosine, the first
specialized Lincoln Log and our first nonstandard nucleoside. Adenosine (A) is
deaminated to form an inosine (I).
There are many functions for inosine editing. Changes from A
to I in mRNA alter the protein made since the inosines get read as G’s. Genomically
coded A’s end up being read as G’s in the mRNA, and this it changes the gene product! We have many more inosine changes
than other primates do.
Many of these A to I edits in humans are related to brain development and are a big reason why we are smarter than chimps.
There is also A to I editing in regulatory RNAs called miRNAs (micro RNA). The miRNAs suppress (prevent)
translation of some transcripts, but editing of the pre-miRNA makes it bind
less well to protein complexes that process the pre- to mature miRNA. More
editing mean less binding of miRNAs, which leads to decreased regulation, more
transcript translation, and increased protein. This may be one way A to I editing increases
human brain power.
Inosine and adenosine accumulate extracellularly during
hypoxia/ischaemia (lack of oxygen or blood flow) in the brain and may act as
neuroprotectants. A new study extends this protective action to the spinal cord in rats in a hypoxic environment. To characterize hypoxia-evoked A and I
accumulation, they examined the effect of hypoxia on the extracellular
levels of adenosine and inosine in isolated spinal cords from rats. "Isolated" means the rats and their spinal cords were not necessarily in the same room at the time - so it could be a while before this helps humans.
But perhaps the most common use for I is to alter tRNA binding
to amino acids and to the target codons. A to I editing can occur in the
anticodon, and change which amino acid is placed in the growing peptide. This
is especially true in many organisms for the amino acid isoleucine. Many tRNAs
will insert an isoleucine into the protein only when the anticodon of the tRNA has been
edited to contain an I in the first position (equivalent to the wobble position
of the mRNA codon).
Other nonstandard (modified) nucleosides also work in tRNAs.
Lysidine, dihydrouridine, and pseudouridine
are some of the more common specialized Lincoln Logs – or maybe we should
stick to calling them nonstandard nucleosides. They can be found in the tRNAs
of organisms from each of the three domains of life (archaea, bacteria, and
eukaryotes). For example, psycrophiles
– organisms that grow at very low temperatures – have 70% more dihydrouridines
because they help the tRNAs to flex as they need to, even at subfreezing
temperatures.
Found mostly in tRNAs, but not exclusively in tRNAs, there
are over 100 non-standard nucleosides. Many times they function to increase
tRNA binding to transcripts via the anticodon-codon, or increase the binding of
the amino acid to the tRNA. They ultimately work to increase translation
efficiency. They are weird and are exceptions, but we can’t live without them.
Next week we can spend some time talking about exceptions in the realm of lipids, the last of our four biomolecules.
For
more information or classroom activities, see:
RNA
editing –